Neoplasia
Editors: Tornetta, Paul; Einhorn, Thomas A.; Damron, Timothy A.
Title: Oncology and Basic Science, 7th Edition
Copyright ©2008 Lippincott Williams & Wilkins
> Table of Contents > Section IV – Basic Science > 28 – Neoplasia
28
Neoplasia
Matthew J. Allen
Neoplasia, literally new growth,
is the result of disturbances in cell growth and/or survival. The
clinical manifestation of this uncontrolled cell growth is the
formation of a tumor. In some cases, for example in the lymph nodes or
skin, the tumor may be palpable as a swelling or lump; in many cases,
however, the tumor develops in an occult fashion and may be identified
only after the patient presents with clinical symptoms. Definitive
diagnosis of the lesion as a neoplasm depends on cytological or
histological examination of a representative biopsy sample. At the same
time, the pathologist will also determine whether the tumor is benign
(unlikely to spread, or metastasize) or malignant (likely to
metastasize). The term cancer, although generally used to describe all tumors, should be restricted to neoplastic lesions with malignant potential.
is the result of disturbances in cell growth and/or survival. The
clinical manifestation of this uncontrolled cell growth is the
formation of a tumor. In some cases, for example in the lymph nodes or
skin, the tumor may be palpable as a swelling or lump; in many cases,
however, the tumor develops in an occult fashion and may be identified
only after the patient presents with clinical symptoms. Definitive
diagnosis of the lesion as a neoplasm depends on cytological or
histological examination of a representative biopsy sample. At the same
time, the pathologist will also determine whether the tumor is benign
(unlikely to spread, or metastasize) or malignant (likely to
metastasize). The term cancer, although generally used to describe all tumors, should be restricted to neoplastic lesions with malignant potential.
Prevalence
-
Epidemiologic data for cancerous
neoplasms, while only the tip of the iceberg in terms of overall
neoplasms, are much more readily available.-
Cancer is the second most common cause of death in the United States (heart disease remains the number-one cause of mortality).
-
According to the American Cancer Society, over 291,000 men and 273,000 women are expected to die as a result of cancer in 2006.
-
Breast and prostate cancers are the most common cancers, but lung cancer remains the most lethal.
-
Overall, the lifetime risk of developing cancer is 1 in 2 for men and women in the United States.
-
Etiology
It is generally accepted that some degree of chromosomal
abnormality is required for cancer to develop. Most cancer cells
display evidence of defects in either chromosome number or composition,
and it is presumed that these genomic changes lie at the heart of the
cancer phenotype. However, with relatively few exceptions, the
mechanisms through which these alterations lead to cancer remain
unclear. Cancer is a disease that develops by clonal expansion from a
single abnormal cell. The underlying abnormality is typically genetic,
but with successive cycles of cell division there is potential for
additive damage through both genetic and nongenetic mechanisms. The end
result of these changes is an expanding clone of cells that is
unresponsive to normal growth controls.
abnormality is required for cancer to develop. Most cancer cells
display evidence of defects in either chromosome number or composition,
and it is presumed that these genomic changes lie at the heart of the
cancer phenotype. However, with relatively few exceptions, the
mechanisms through which these alterations lead to cancer remain
unclear. Cancer is a disease that develops by clonal expansion from a
single abnormal cell. The underlying abnormality is typically genetic,
but with successive cycles of cell division there is potential for
additive damage through both genetic and nongenetic mechanisms. The end
result of these changes is an expanding clone of cells that is
unresponsive to normal growth controls.
Loss of Normal Growth Control in Cancer Cells
Normal cellular growth is controlled by four major
groups of regulatory proteins. Changes in the level of expression or in
the genetic sequence of these regulatory proteins have the capacity to
alter their function and, as a result, disturb normal cellular growth.
groups of regulatory proteins. Changes in the level of expression or in
the genetic sequence of these regulatory proteins have the capacity to
alter their function and, as a result, disturb normal cellular growth.
-
Major groups of regulatory proteins are involved in normal cellular growth control.
-
Growth factors
-
Definition: circulating factors or local factors that act on cells with specific receptors
-
Mechanisms of disturbance: Inappropriate expression (up- or down-regulation) can lead to uncontrolled cell growth.
-
Example: Disturbances in fibroblast growth factor (FGF) have been implicated in the development of sarcomas.
-
P.526 -
-
Growth factor receptors
-
Definition: Typically expressed either on
the cell surface or within the cytoplasm, these receptors bind and
transduce external signals from growth factors into intracellular
signals. -
Mechanisms of disturbance: Disturbances
in receptor activity may increase or decrease receptor numbers on the
cell, or they may involve dysregulated receptors that are permanently
“on,” even in the absence of the growth factor signal.-
Example: Overexpression of c-erb2/HER2 is implicated in breast cancer.
-
-
-
Intracellular signaling proteins
-
Definition: These proteins transfer the
signal from surface receptors to the nucleus, where the target is most
often a nuclear transcription factor (see below). -
Mechanisms of disturbance: Overactivity
of these signaling proteins, for example as a result of constitutive
overexpression of the gene, leads to uncontrolled nuclear signaling and
activation of downstream genes, many of which regulate cellular growth
and/or survival.-
Example: Overexpression of the ras family of oncogenes is found in many tumors.
-
-
-
Nuclear transcription factors
-
Definition: These control elements act directly on DNA, resulting in gene transcription.
-
Mechanisms of disturbance: In some cases
chromosomal damage directly affects the expression of the transcription
factor, while in others the effects are mediated via changes in the
expression of tumor suppressor proteins.-
Examples: A number of transcription
factors have been implicated in human cancer, including E2F (involved
in the cell cycle), p53, and MDM2 (see below).
-
-
-
-
Downstream effects of mutations in any of these control elements
-
Depend on the nature of the affected gene and the extent of the change
-
Defects in one copy of a tumor suppressor gene may have no clinical consequence as long as the second copy is normal.
-
Defects in the androgen receptor may be silent in females but cause feminization in males.
-
In the most extreme cases, defects in just one copy of a gene may still lead to clinical disease.
-
Example: autosomal-dominant conditions such as achondroplasia (associated with a defective gene for the FGF3 receptor)
-
-
-
Genetic Alterations in Cancer
Mutations and Cancer
-
Definition of mutation: changes in the normal sequence of DNA as a result of defects in DNA replication
-
Causes of mutations
-
Endogenous defects in DNA replication and/or DNA repair
-
Exogenous factors such as radiation or chemical carcinogens
-
-
Timing of mutations relative to cell division
-
Only at the time of DNA replication is damage converted into a change in the DNA sequence.
-
Nondividing cells are less susceptible to
mutation than are cells that are highly proliferative (e.g.,
hematopoietic tissues, skin, intestinal tract).
-
-
Targets for genetic mutation in cancer
-
Typically regulatory elements that affect cellular growth, differentiation, and survival (see above)
-
-
Clinical importance of mutations
-
Associations between a particular cancer and a specific pattern of mutation exist.
-
Associations are rarely absolute.
-
Used in some cases to confirm histological diagnoses
-
Example: 11:22 translocation in Ewing sarcoma
-
-
-
It is common to find that an individual
tumor contains multiple gene defects, and it is the combination of
defects, rather than any single mutation, that determines the phenotype
of the tumor in terms of growth, risk of metastasis, and response to
therapy.-
Examples: conventional osteosarcoma, chondrosarcoma
-
-
Cancer and the Cell Cycle
As described in Chapter 13, Cellular and Molecular Biology,
the cell cycle is regulated by a complex series of control elements
that ensure the integrity of the DNA, the accuracy of DNA replication,
and the integrity of the cytoskeletal elements that facilitate the
division of the two sets of DNA into the daughter cells. Disturbances
in one or more of these control elements can lead to a spectrum of
problems, including decreased proliferative capacity, abnormal cellular
differentiation, resistance to apoptosis, or unchecked cellular
proliferation. It is therefore not surprising that disturbances in the
normal cell cycle lie at the heart of many human cancers.
the cell cycle is regulated by a complex series of control elements
that ensure the integrity of the DNA, the accuracy of DNA replication,
and the integrity of the cytoskeletal elements that facilitate the
division of the two sets of DNA into the daughter cells. Disturbances
in one or more of these control elements can lead to a spectrum of
problems, including decreased proliferative capacity, abnormal cellular
differentiation, resistance to apoptosis, or unchecked cellular
proliferation. It is therefore not surprising that disturbances in the
normal cell cycle lie at the heart of many human cancers.
Cyclins and Cyclin-Dependent Kinases (Cdks) (Fig. 28-1)
-
Critical role in controlling the orderly progression of cells through the cell cycle
-
Mammalian species have at least 10 cyclins (cyclins A, B, C, etc.).
-
Cyclins cannot act alone.
-
Require activation by binding of a cyclin-dependent kinase (Cdk) to form Cdk—cyclin complexP.527
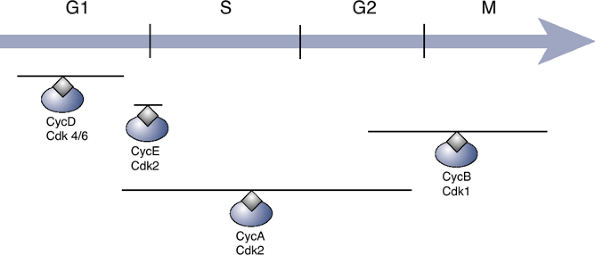 Figure 28-1
Figure 28-1
Regulation of the cell cycle by cyclins and cyclin-dependent kinases.
The gray lines above each complex indicate the stages of the cell cycle
during which they are most active.-
In mammalian species there are at least six Cdks (Cdk 1 through 6).
-
-
Subsequent phosphorylation of the Cdk within the Cdk—cyclin complex
-
Phosphorylating enzyme known as Cdk-activating kinase (CAK)
-
-
Sequence in mammalian cells
-
Cell cycle initiated by binding of cyclin D to either Cdk4 or Cdk6
-
Final phase catalyzed by binding between cyclin B and Cdk1 to produce a complex known as mitosis-promoting factor (MPF)
-
-
-
Control over cyclin—Cdk activity exerted through effects at multiple levels
-
Cyclin synthesis or breakdown
-
Cyclin—Cdk binding
-
CAK activity
-
Direct inhibition of Cdk activity by Cdk inhibitors
-
Proto-Oncogenes, Cellular Oncogenes, and Viral Oncogenes
-
Oncogenes
-
DNA sequences that regulate the growth of tumor cells
-
Derived from proto-oncogenes
-
-
Proto-oncogenes
-
DNA sequences in the healthy cell that
encode transcription factors, intracellular signaling pathways, and
receptors that control normal cellular growth, differentiation, and
apoptosis -
Importance of these genes as regulatory elements
-
Highlighted by the fact that they have been highly conserved throughout the course of evolution
-
Also strong homology between proto-oncogenes in different vertebrate species and even nonvertebrates (e.g., Drosophila, yeast species)
-
-
-
Conversion of a proto-oncogene into an
oncogene: This key step in cancer initiation involves activation
through one of three mechanisms:-
Point mutation: Changes in the nucleic acid sequence can render the gene product resistant to inhibitory elements.
-
Translocation: If the oncogene becomes
associated with a new promoter, it may be expressed at abnormally high
levels. Alternatively, combination of the oncogene with a gene sequence
can lead to the formation of a fusion gene whose product can enhance
cellular growth. -
Amplification: production of multiple copies of the oncogene
-
-
Specific important oncogenes (Table 28-1)
-
Many originally described in retroviruses known to be associated with neoplastic diseases in animals
-
Rous sarcoma virus
-
Avian erythroblastosis virus
-
-
Thought that these oncogenes were introduced into the viral genome when the viral DNA became integrated into the cellular DNA
-
Viral oncogene is capable of inducing
cellular transformation directly because it has already been activated,
either by mutations within its sequence or by the acquisition of
multiple copies of the oncogene within the viral genome.
-
-
Tumor Suppressor Genes
Tumor suppressor genes encode proteins that regulate
cell growth. Inhibition of tumor suppressor activity is associated with
increased cell growth, genomic instability, and an increased
susceptibility to cancer.
cell growth. Inhibition of tumor suppressor activity is associated with
increased cell growth, genomic instability, and an increased
susceptibility to cancer.
p53
-
Normal role of p53 in healthy cells
-
Activated by DNA damage or hypoxiaP.528Table 28-1 Cellular Oncogenes and Their Association with Human Cancer
Oncogene Function Association with Human Cancer erb-B1 Receptor for epidermal growth factor (EGF) Bladder, lung, colon, breast carcinoma erb-B2 (also known as Her2/neu) Tyrosine kinase Breast, ovarian, lung, pancreatic carcinoma Sis Growth factor (PDGF) Non–small cell lung cancer Src Tyrosine kinase Breast, pancreatic, colon carcinoma ras family G-protein Colon, pancreas, lung, bladder carcinoma bcl1 Cyclin D1 B-cell malignancies -
Protein product with multitude of diverse functions, perhaps best summarized as “guardian of the genome”
-
Functions as a transcription factor, regulating downstream genes involved in cell cycle arrest, DNA repair, and apoptosis
-
Major gene product induced by p5 3 is WAF1/CIP1/p21, inhibitor of Cdk-2
-
p21 activation blocks progression into the S phase.
-
-
-
-
Dysfunction of p53 tumor suppressor gene
-
Mutations in p53 have been found in approximately 50% of all human cancers (Table 28-2).
-
Loss of p53 function induces genomic
instability, impaired apoptosis, and diminished cell cycle restraint,
all of which are critical elements favoring malignancy. -
Loss of p53 activity confers resistance
to apoptosis and, as a result, decreases the sensitivity of tumor cells
to radiation and chemotherapy.
-
Retinoblastoma (Rb)
-
Normal role of Rb in healthy cells:
inactivates cyclin—Cdk complexes that are required for transition
through the G1/S checkpoint -
Dysfunction of Rb tumor suppressor gene:
Initially described in the hereditary form of retinoblastoma, the Rb
oncogene has subsequently been implicated in a variety of human cancers
(see Table 28-2), including osteosar
MDM2
-
Human homolog of the murine double minute 2 gene (MDM2), first identified as an amplified gene in a transformed mouse cell line
-
Normal role of MDM2 in healthy cellsTable 28-2 Tumor Suppressor Genes and Their Association with Human Cancer
Gene Product Mechanism of Action Association with Human Cancer p53 Regulates cell cycle progression and apoptosis Colon, lung, breast, esophagus, brain, liver; leukemia; osteosarcoma and rhabdomyosarcoma Rb Inactivates cyclin–Cdk complexes and stops cell cycle progression Lung, bladder, breast carcinoma; osteosarcoma MDM2 Downregulates p53 activity Soft tissue sarcomas, metastatic osteosarcoma BRCA1 Ubiquitin ligase activity Early-onset breast and ovarian carcinoma P.529-
Controls the activity of p53 via two distinct mechanisms
-
MDM2 binds to p53 and physically prevents it from activating target DNA sequences
-
MDM2 transfers a ubiquitin tag to p53, thereby marking it for degradation by the ubiquitin—proteasome system
-
-
In response to DNA damage, p53 can activate MDM2 expression.
-
Since MDM2 inhibits p53 activity, negative feedback control loop is established.
-
-
-
Dysfunction of MDM2: Overexpression of MDM2 leads to functional inactivation of p53.
-
Amplification of MDM2 has been reported in over one third of soft tissue sarcomas, as well as in a number of other cancers (see Table 28-2).
-
Even in the face of normal p53, constitutive overexpression of MDM2 appears to increase the risk of neoplastic transformation.
-
Nongenetic Alterations in Cancer
Although the vast majority of cancer research focuses on
the role of chromosomal mutations, it is also possible to see changes
in cell proliferation and differentiation in the absence of permanent
changes in the DNA. Common examples would include tumor viruses and the
hormonal environment to which tissues are exposed.
the role of chromosomal mutations, it is also possible to see changes
in cell proliferation and differentiation in the absence of permanent
changes in the DNA. Common examples would include tumor viruses and the
hormonal environment to which tissues are exposed.
Tumor Viruses
-
Both RNA viruses and DNA viruses have been implicated in cancer.
-
Most of the information on RNA tumor
viruses comes from naturally occurring diseases in animals (e.g., Rous
sarcoma virus, avian erythroblastosis virus).-
With the exception of human T-cell
leukemia virus (HTLV-1), there does not appear to be a strong
association between RNA viruses and cancer in humans.
-
-
Of the DNA viruses that are linked to cancer, the most important associations appear to be between:
-
Epstein-Barr and Burkitt’s lymphoma/nasopharyngeal carcinoma
-
Human papillomavirus and cervical cancer
-
Hepatitis B and hepatocellular carcinoma
-
Hormonal Factors
-
Estrogen and cancer
-
Prolonged exposure to estrogens is a risk factor in the development of both breast and endometrial cancer.
-
Most of the evidence points toward estrogens as promoters rather than initiators of cancer.
-
By increasing the proliferative rate within target tissues, they increase the likelihood of developing genomic instability.
-
-
-
Approximately 70% of primary breast cancers express the estrogen receptor and are responsive to circulating estrogen.
-
-
Androgens and prostate cancer
-
Androgens stimulate the growth of prostate cancer cells in vitro and in vivo.
-
-
Hormonal-based therapies: Strategies that
reduce endogenous hormone synthesis (e.g., by surgical or chemical
ablation) or that interfere with receptor activation (e.g., selective
estrogen receptor modifiers [SERMs]) are effective in the management of
primary breast or prostate cancer.
Chemical and Physical Agents as Causes of Cancer
Many of the early studies on the etiology of cancer
focused on environmental factors such as chemical and physical stimuli.
Classic studies included those of Sir Percival Potts on skin cancer in
chimney sweeps, the identification of bone cancer in women exposed to
radium during the painting of watch faces, and the observation of a
causal link between radiation exposure and leukemia. These studies led
to the general concept of DNA as the target for damage in cancer, as
well as to the concepts of promoters and initiators in the pathogenesis
of cancer. The list of chemical/physical agents that are linked to
cancer continues to grow. However, with the exception of the
association between ionizing radiation and osteosarcoma, none appears
to play a significant role in the development of musculoskeletal
neoplasms.
focused on environmental factors such as chemical and physical stimuli.
Classic studies included those of Sir Percival Potts on skin cancer in
chimney sweeps, the identification of bone cancer in women exposed to
radium during the painting of watch faces, and the observation of a
causal link between radiation exposure and leukemia. These studies led
to the general concept of DNA as the target for damage in cancer, as
well as to the concepts of promoters and initiators in the pathogenesis
of cancer. The list of chemical/physical agents that are linked to
cancer continues to grow. However, with the exception of the
association between ionizing radiation and osteosarcoma, none appears
to play a significant role in the development of musculoskeletal
neoplasms.
Identifying and Classifying Chromosomal Abnormalities in Cancer
Normal Human Genome and Cytogenetic Analysis
The normal human genome consists of 46 chromosomes: 22
pairs of autosomal chromosomes (one copy from each parent) and one pair
of sex chromosomes (XY or XX). The identification of chromosomal
abnormalities involves cytogenetic analysis. Traditionally this was
performed by visual examination of the chromosomes in a so-called
metaphase spread (the staining of chromosomes is most easily
accomplished when cells are in the metaphase) and identification of
gross abnormalities in either chromosome number (e.g., three copies of
chromosome 21 in Down syndrome), size (e.g., abnormally short or long
chromosomes are indicative of translocations), or staining pattern.
However, these time-consuming techniques have now been revolutionized
by the development of molecular techniques such as fluorescent in situ
hybridization (FISH), in which fluorescent molecular probes are used to
identify the location of specific sequences within the chromosomes.
pairs of autosomal chromosomes (one copy from each parent) and one pair
of sex chromosomes (XY or XX). The identification of chromosomal
abnormalities involves cytogenetic analysis. Traditionally this was
performed by visual examination of the chromosomes in a so-called
metaphase spread (the staining of chromosomes is most easily
accomplished when cells are in the metaphase) and identification of
gross abnormalities in either chromosome number (e.g., three copies of
chromosome 21 in Down syndrome), size (e.g., abnormally short or long
chromosomes are indicative of translocations), or staining pattern.
However, these time-consuming techniques have now been revolutionized
by the development of molecular techniques such as fluorescent in situ
hybridization (FISH), in which fluorescent molecular probes are used to
identify the location of specific sequences within the chromosomes.
Terminology Describing Normal and Abnormal Chromosomal Arrangements
-
Chromosomes are identified by a number or, in the case of the sex chromosomes, a letter.
-
Each chromosome is divided into arms:
-
Short arm = designated as p
-
Long arm = q
-
-
Each arm is divided into regions.
-
Each region is divided into bands.
-
Both regions and bands are numbered, with
the numbers increasing with increasing distance from the middle of the
chromosome (the centromere).
-
-
Summary: CARB (Chromosome, Arm, Region, Band)
-
Accordingly, 13q 13 refers to the third band on the first region on the long arm of chromosome 13.
-
P.530
Disturbances in the Normal Arrangement of Chromosomes
-
Four mechanisms: translocation, point mutation, deletion, amplification
-
Most common mechanism associated with
malignancy is translocation, the result of two chromosomes breaking and
sequences from one becoming incorporated into the other (Fig. 28-2).-
Effects depend on the nature of the sequence that moves and on the location into which it moves.
-
Clinically important examples of chromosomal translocations are summarized in Table 28-3.
-
Chromosomal Rearrangements in Sarcoma
As can be seen in Table 28-3, a
number of translocations have been identified in sarcomas. The strong
association between these rearrangements and the clinical tumor makes
the identification of these translocations useful from a diagnostic
perspective. The utility of this type of molecular diagnostic approach
is such that it is becoming an integral part of the pathological
confirmation and classification of sarcomas. Some of the specific
sarcomas and their chromosomal rearrangements are reviewed below.
number of translocations have been identified in sarcomas. The strong
association between these rearrangements and the clinical tumor makes
the identification of these translocations useful from a diagnostic
perspective. The utility of this type of molecular diagnostic approach
is such that it is becoming an integral part of the pathological
confirmation and classification of sarcomas. Some of the specific
sarcomas and their chromosomal rearrangements are reviewed below.
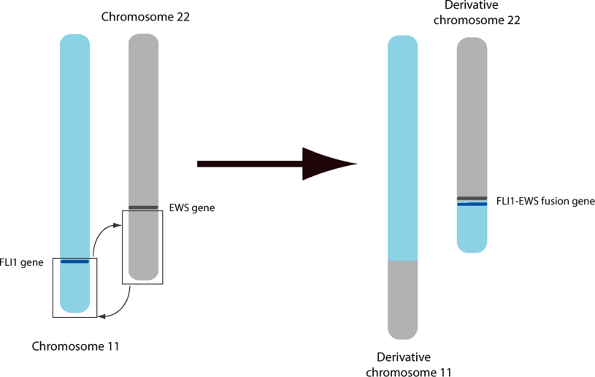
Ewing Sarcoma/Primitive Neuroectodermal Tumor (PNET)
-
Products of t(11:22)
-
EWS/FLI1 fusion gene: over 90% of Ewing sarcoma/PNET lesions
-
EWS/ERG and EWS/ETV1 genes: ~5% of cases each
-
-
Individual components
![]() Figure 28-2
Figure 28-2
Terminology of chromosomal rearrangements. The chromosomes that are
affected in the translocation are indicated in the first grouping, in
parentheses, while the region/band information is contained in the
second grouping, in square brackets. The entire sequence is preceded by
the letter t, indicating a translocation. This particular rearrangement is seen in Ewing sarcoma.P.531Table 28-3 Chromosomal Translocations Associated with Specific Human Musculoskeletal Cancers and Chronic Myeloid LeukemiaDisease Translocation(s) Product(s) Chronic myeloid leukemia t(9:22) (q34:q11) (Philadelphia chromosome) BCR-ABL Ewing sarcoma/primitive neuroectodermal tumors (PNET) t(11:22) (q24:q12) EWS-FLI1
EWS-ERG
EWS-ETV1Synovial sarcoma t(x:18) (p11:q11) SYT-SSX Desmoplastic/small round cell sarcoma t(11:22) (q13:q12) EWS-WT1 Myxoid/round cell liposarcoma t(12:16) (q13:p11) CHOP-TLS/FUS t(12:22) (q13:q12) CHOP-EWS Myxoid chondrosarcoma t(9:22) (q31:q12) EWS-TEC Alveolar rhabdomyosarcoma t(2:13) (q35–37:q14) PAX3-FKHR -
EWS gene: encodes a protein that binds RNA
-
FLI1, ERG, and ETV1: all members of the ETS family of transcription factors
-
-
Evidence of importance
-
EWS-FLI1 gene is capable of transforming cells in vitro.
-
Downregulation of EWS-FLI1 activity suppresses the growth of Ewing sarcoma cells in vitro.
-
Synovial Sarcoma
-
Products of t(x: 18)
-
SYT gene on X chromosome translocates to involve one of two SSX genes on chromosome 18.
-
Present in nearly all synovial sarcomas
-
-
Individual components: SSX1 and SSX2 are DNA-binding proteins that regulate gene transcription.
-
Clinical relevance: The presence of SSX1 rather than SSX2 in the fusion gene appears to have prognostic importance.
-
5-year survival for SYT-SSX2 is 48%, compared with 24% for patients with the SYT-SSX1 translocation.
-
Myxoid/Round Cell Liposarcoma
-
Products of t(12:16)
-
Fuses the CHOP gene on chromosome 12 with the TLS/FUS gene on chromosome 16
-
Present in over three quarters of myxoid/round cell liposarcoma lesions
-
-
Individual components
-
CHOP gene encodes a DNA transcription factor.
-
TLS encodes an activated transcription factor.
-
Targets for these transcription factors are not currently known.
-
Factors Controlling the Growth of Tumors
-
Vascularization
-
Vascularization is required for tumors >2 mm in diameter.
-
Antiangiogenic strategies (e.g., endostatin) show promise in retarding tumor growth.
-
-
pH
-
Oxygen tension
-
Local environment within/around many tumors can be hypoxic.
-
Hypoxia can decrease the effectiveness of standard cancer treatments such as chemotherapy or radiation therapy.
-
-
Endocrine, paracrine, and autocrine factors
-
Estrogen and androgen responsiveness of many breast and prostate cancers (see above)
-
Interleukin (IL)-6, released from bone marrow cells, is known to be a potent mitogen for multiple myeloma.
-
Transforming growth factor beta (TGF-β),
insulin-like growth factor I (IGF-I), and IL-8 have all been implicated
as factors that upregulate the growth of cancer cells within the
skeleton.
-
Tumor Metastasis
Spread of the tumor away from its site of origin is
known as metastasis. In most cancer patients, death results from the
effects of the metastatic tumor rather than the primary tumor per se.
What, then, is it that makes some tumors metastasize? The barriers to
metastasis are immense; the tumor must first enter the systemic
circulation (either vascular
known as metastasis. In most cancer patients, death results from the
effects of the metastatic tumor rather than the primary tumor per se.
What, then, is it that makes some tumors metastasize? The barriers to
metastasis are immense; the tumor must first enter the systemic
circulation (either vascular
P.532
or lymphatic), then travel to target organs, exit the circulation, and then establish a new colony (Fig. 28-3).
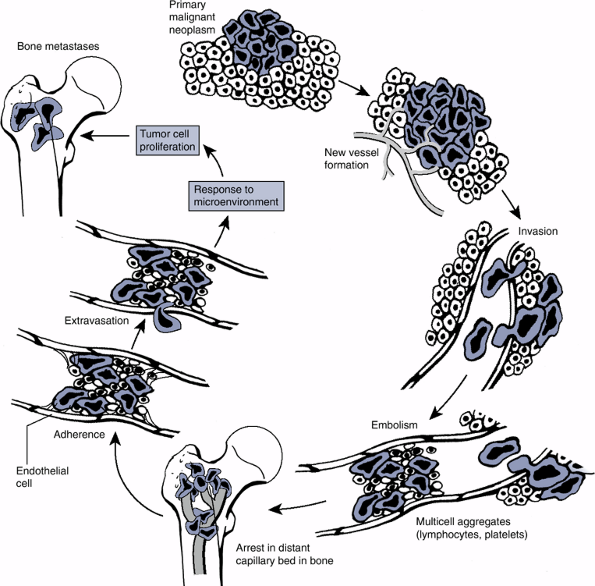 |
|
Figure 28-3
Steps involved in tumor metastasis. (Adapted by permission from Macmillan Publishers Ltd: Nature Reviews: Cancer. Mundy GR. Metastasis to bone: Causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002;2:584–593.) |
Many solid tumors display predictable patterns of
metastasis (e.g., bone, liver, and lung metastasis for many
carcinomas), suggesting that the process of metastasis is far from
random. A number of factors are now known to be important in
controlling tumor cell metastasis (see Chapter 8: Metastatic Disease).
Of particular interest within the context of musculoskeletal neoplasms
are the matrix metallo-proteinases (MMPs), which were discussed in Chapter 13. The clinical consequences of primary and secondary tumors on the skeleton were reviewed in Chapter 8, Metastatic Disease.
metastasis (e.g., bone, liver, and lung metastasis for many
carcinomas), suggesting that the process of metastasis is far from
random. A number of factors are now known to be important in
controlling tumor cell metastasis (see Chapter 8: Metastatic Disease).
Of particular interest within the context of musculoskeletal neoplasms
are the matrix metallo-proteinases (MMPs), which were discussed in Chapter 13. The clinical consequences of primary and secondary tumors on the skeleton were reviewed in Chapter 8, Metastatic Disease.
Suggested Reading
Antonescu C R. The role of genetic testing in soft tissue sarcoma. Histopathology 2006;48(1):13–21.
Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003;3(6):537–549.
Lazar A, Abruzzo LV, Pollock RE, et al. Molecular diagnosis of sarcomas: chromosomal translocations in sarcomas. Arch Pathol Lab Med 2006;130(8):1199–1207.
Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002;2(8):584–593.
Schwartz GK, Shah MA. Targeting the cell cycle: a new approach to cancer therapy. J Clin Oncol 2005;23(36):9408–9421.
Steele RJ, Thompson AM, Hall PA, et al. The p53 tumour suppressor gene. Br J Surg 1998;85(11): 1460–1467.